Remember back in high school biology when you studied genetics and learned about DNA, nucleotides and gene sequencing? Join us for this week’s Tool Talk as we clear away the cobwebs on gene sequencing and learn how ACER scientists are using this process to study the microbial community composition.
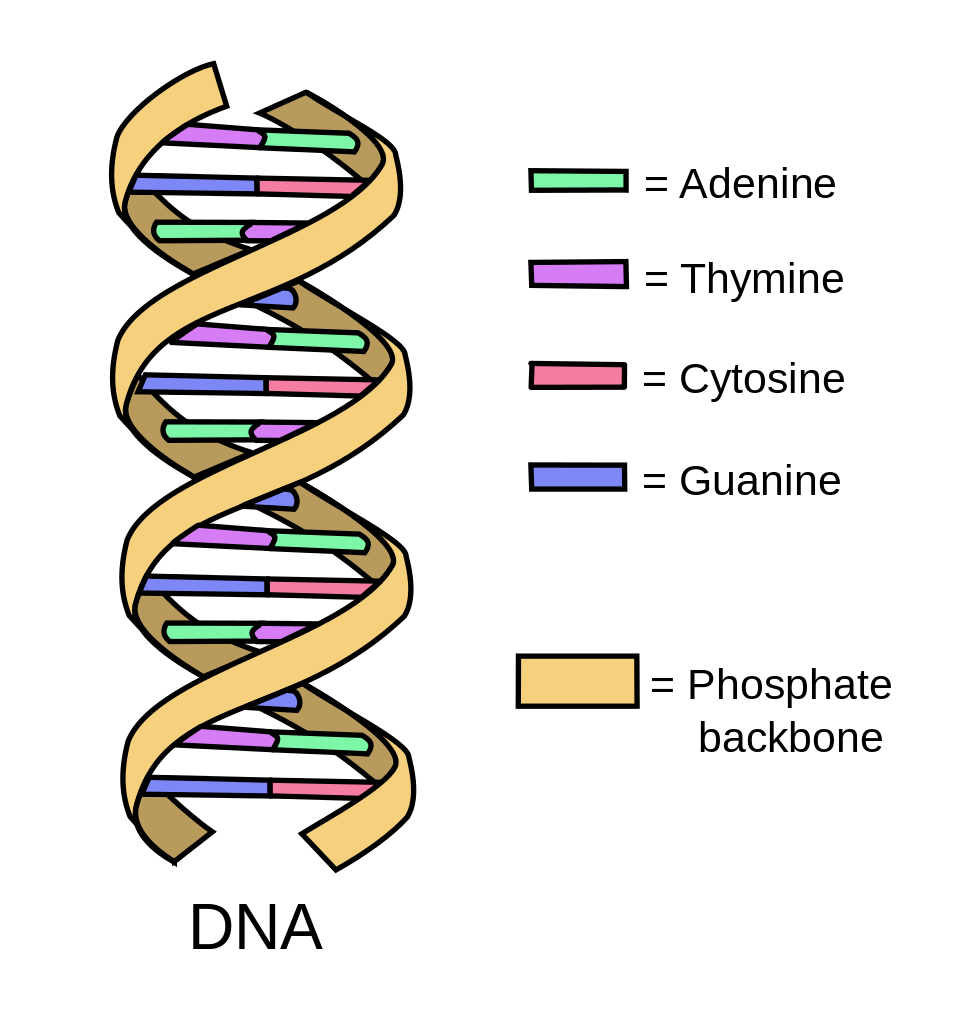
Deoxyribonucleic acid (DNA) is a double helix strand of paired nucleotides that provide the code for building and maintaining an organism. There are four nucleotides or chemical bases (adenine (A), guanine (G) cytosine (C) and thymine (T)) that make up the entire DNA sequence, which for a human is about 3 million bases. The order in which these base pairs sit along the double helix determines an organism’s genes and the process of determining this order is called DNA or gene sequencing.
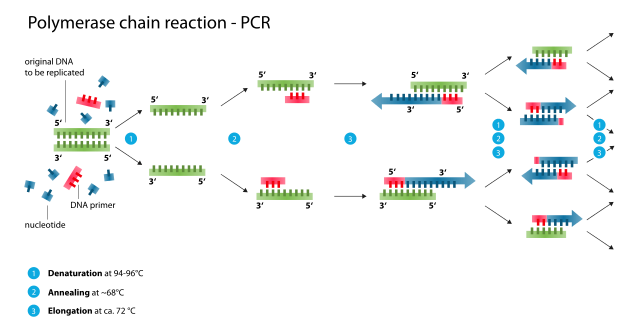
There are several steps involved in sequencing DNA. First, the DNA has to be extracted out of a cell. Next, the DNA strand is broken into fragments to be copied. The copies then undergo a process which separates the DNA fragments by size for identification. One of the main methods scientists use to sequence DNA is called the polymerase chain reaction (PCR).
For this process, a single copy of DNA, one of interest to the researcher, is amplified through a processes of heating (to denature the DNA protein structure so it can be copied) and cooling (to promote DNA replication) to create thousands to millions of copies of that particular DNA sequence. Polymerase is an enzyme that helps to elongate the strand of DNA along with primers, which are short strands of DNA sequences that are complimentary to the DNA segments being amplified.
The next step after PCR in gene sequencing is electrophoresis. In this process the DNA products from PCR are placed in a gel and an electric current is applied. This separates the DNA fragments by length (i.e. the number of base pairs) with shorter lengths moving faster and further through the pores of the gel. The nucleotide bases are then identified by a dye and the information sent into a computer where the sequence can be analyzed.
ACER scientists are using gene sequencing to identify and compare bacteria present within sediment samples. Bacteria, which we introduced in our Word Wednesday series, are prokaryotic and have a single, circular and naked DNA molecule. They also have a section of DNA known as 16s ribosomal RNA (rRNA) which is found within most bacteria and archaea and therefore used for bacterial gene sequencing. Once the DNA has been amplified the fragment DNA sequence can be entered into a 16s rRNA databases which allows scientists to analyze their DNA segments against known sequences to identify and compare bacteria.
What can we learn from gene sequencing bacteria? Currently the effects of oiling in nearshore sediments on the microbial community structure and denitrification rates remain unknown. By identifying bacterial species at locations that received varying levels of oiling, ACER scientists may be able to understand how the oiling affected the denitrifying microbial community in Louisiana salt marshes.